Rapid cost-effective viral genome sequencing by V-seq
By Longhua Guo, James Boocock, ... Joshua S. Bloom, Leonid Kruglyak, Yi Yin | Preprint
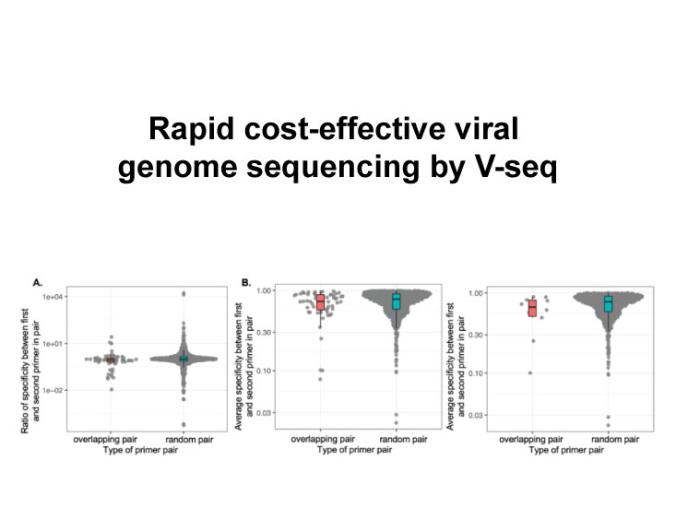
Conventional methods for viral genome sequencing largely use metatranscriptomic approaches or, alternatively, enrich for viral genomes by amplicon sequencing with virus-specific PCR or hybridization-based capture. These existing methods are costly, require extensive sample handling time, and have limited throughput. Here, we describe V-seq, an inexpensive, fast, and scalable method that performs targeted viral genome sequencing by multiplexing virus-specific primers at the cDNA synthesis step. We designed densely tiled reverse transcription (RT) primers across the SARS-CoV-2 genome, with a subset of hexamers at the 3’ end to minimize mis-priming from the abundant human rRNA repeats and human RNA PolII transcriptome. We found that overlapping RT primers do not interfere, but rather act in concert to improve viral genome coverage in samples with low viral load. We provide a path to optimize V-seq with SARS-CoV-2 as an example. We anticipate that V-seq can be applied to investigate genome evolution and track outbreaks of RNA viruses in a cost-effective manner. More broadly, the multiplexed RT approach by V-seq can be generalized to other applications of targeted RNA sequencing.
By Longhua Guo, James Boocock, Jacob M. Tome, Sukantha Chandrasekaran, Evann E. Hilt, Yi Zhang, Laila Sathe, Xinmin Li, Chongyuan Luo, Sriram Kosuri, Jay A. Shendure, Valerie A. Arboleda, Jonathan Flint, Eleazar Eskin, Omai B. Garner, Shangxin Yang, Joshua S. Bloom, Leonid Kruglyak, Yi Yin
